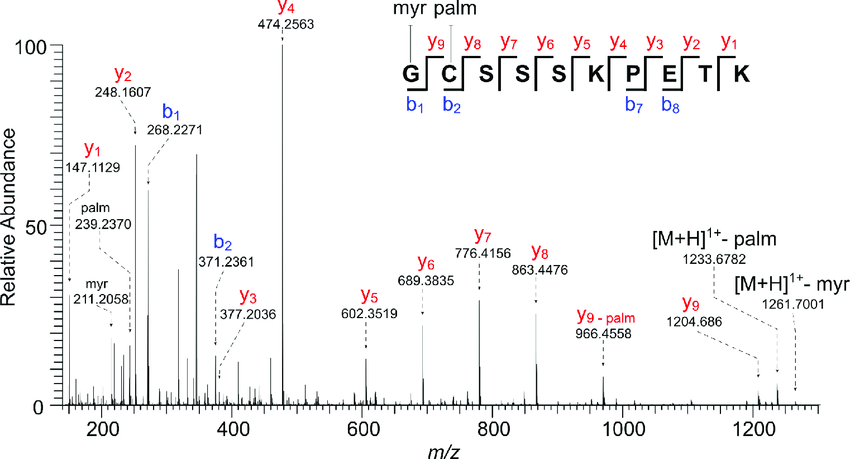
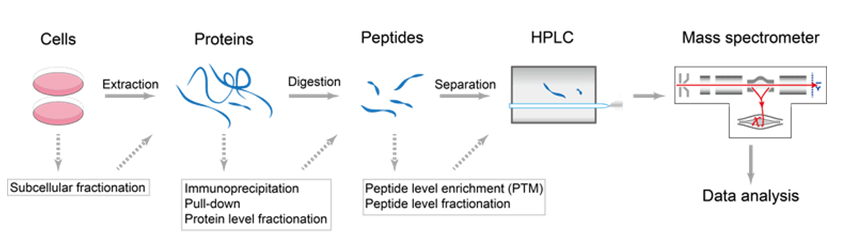
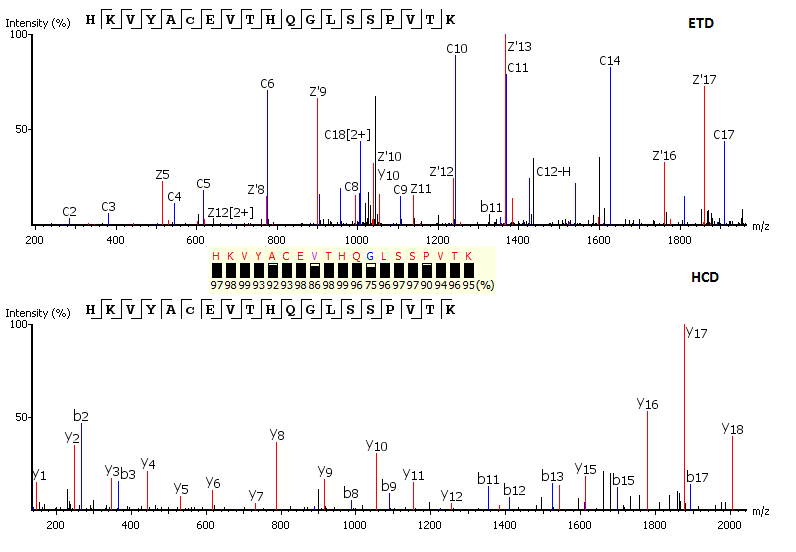
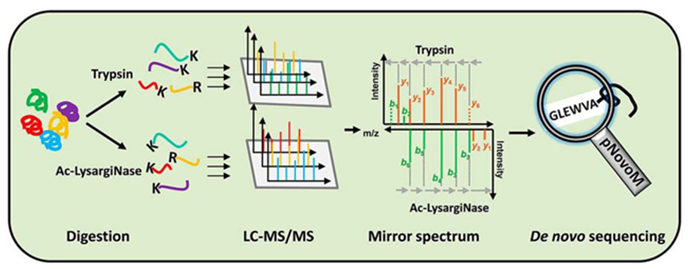
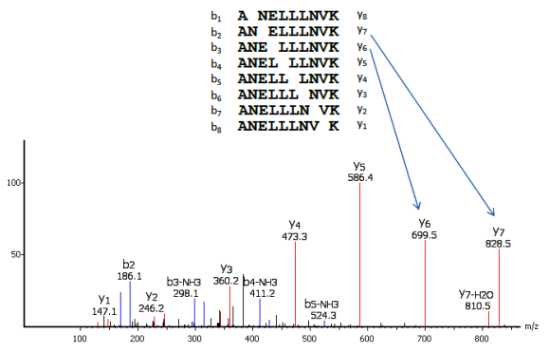
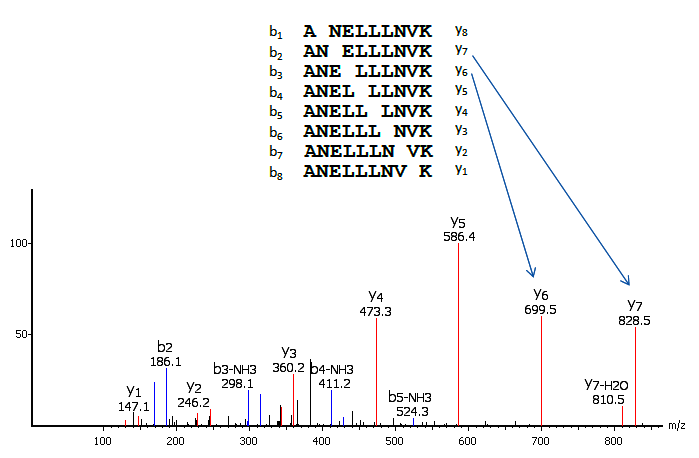
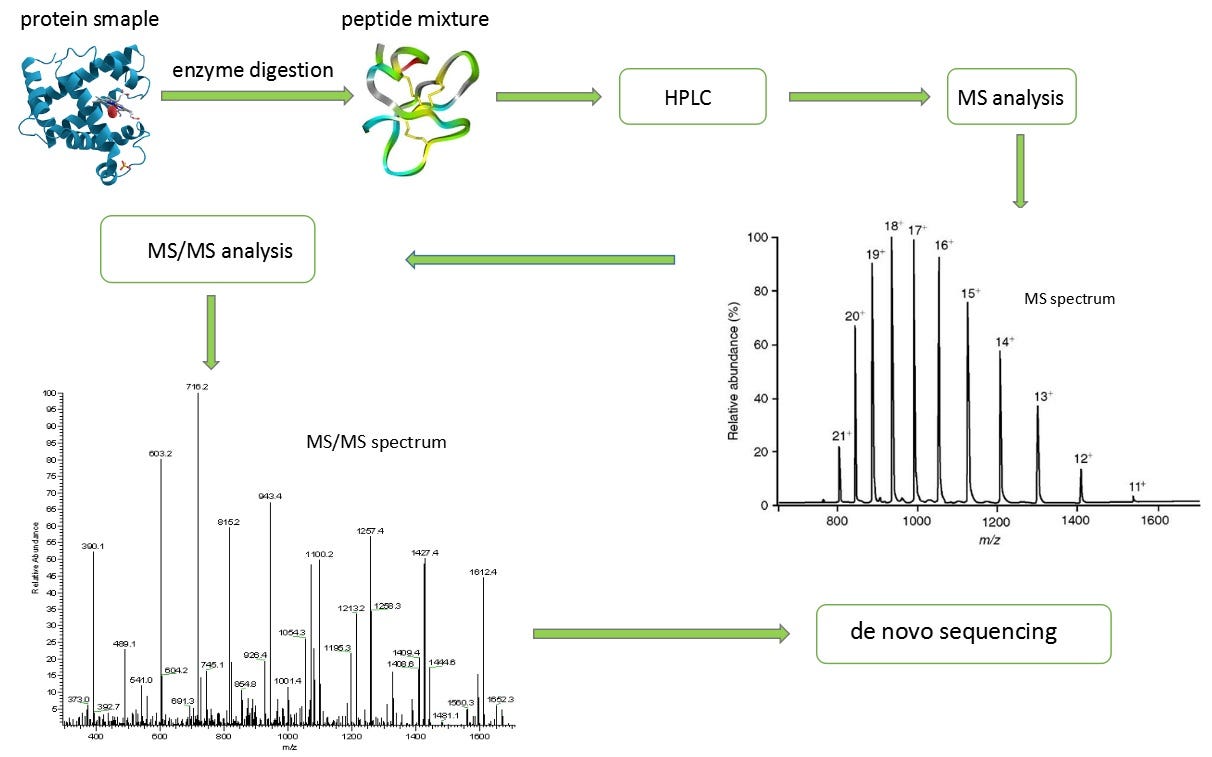
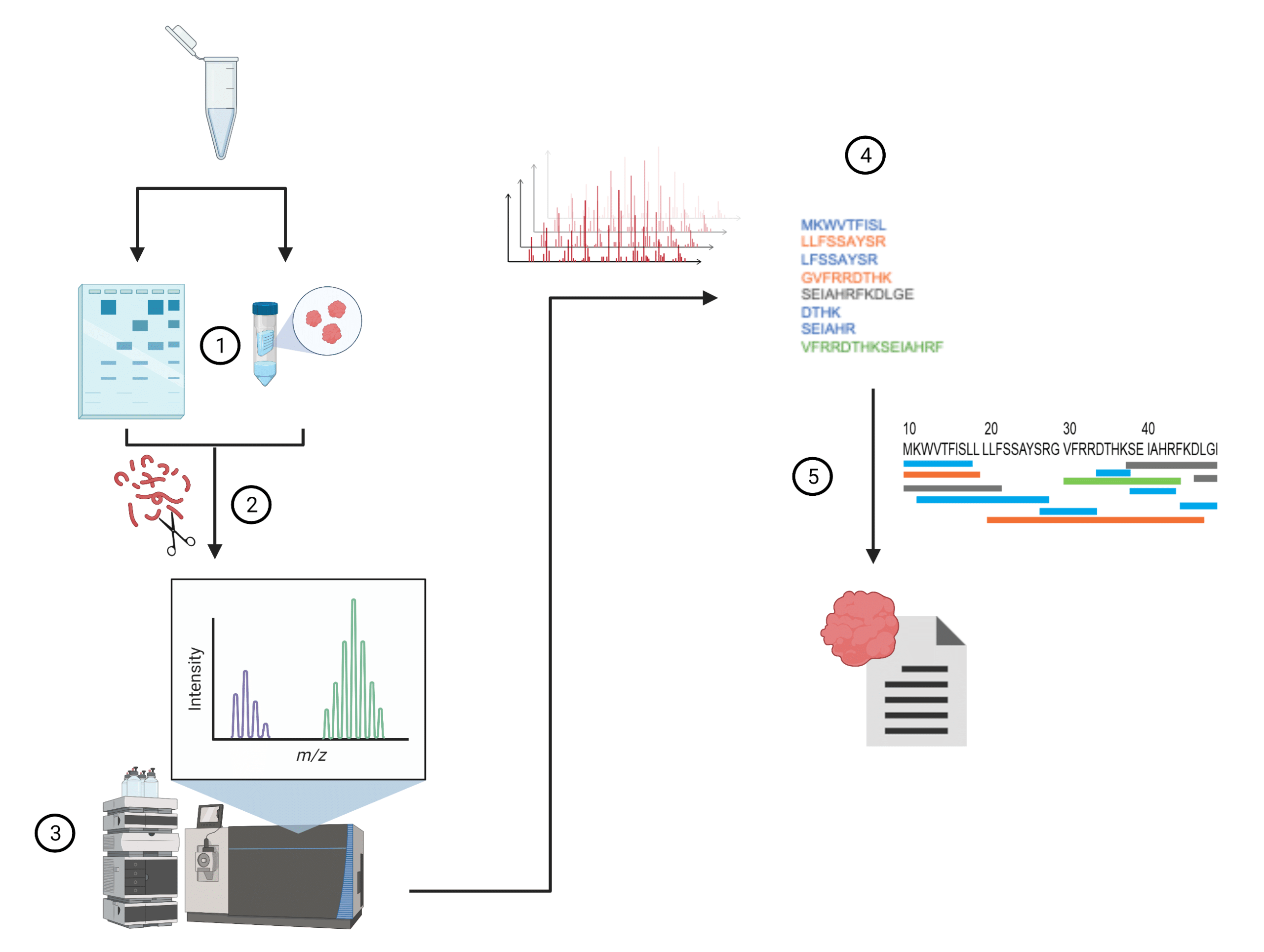
Precision De Novo Peptide Sequencing Using Mirror Proteases of Ac-LysargiNase and Trypsin for Large-scale Proteomics - ScienceDirect

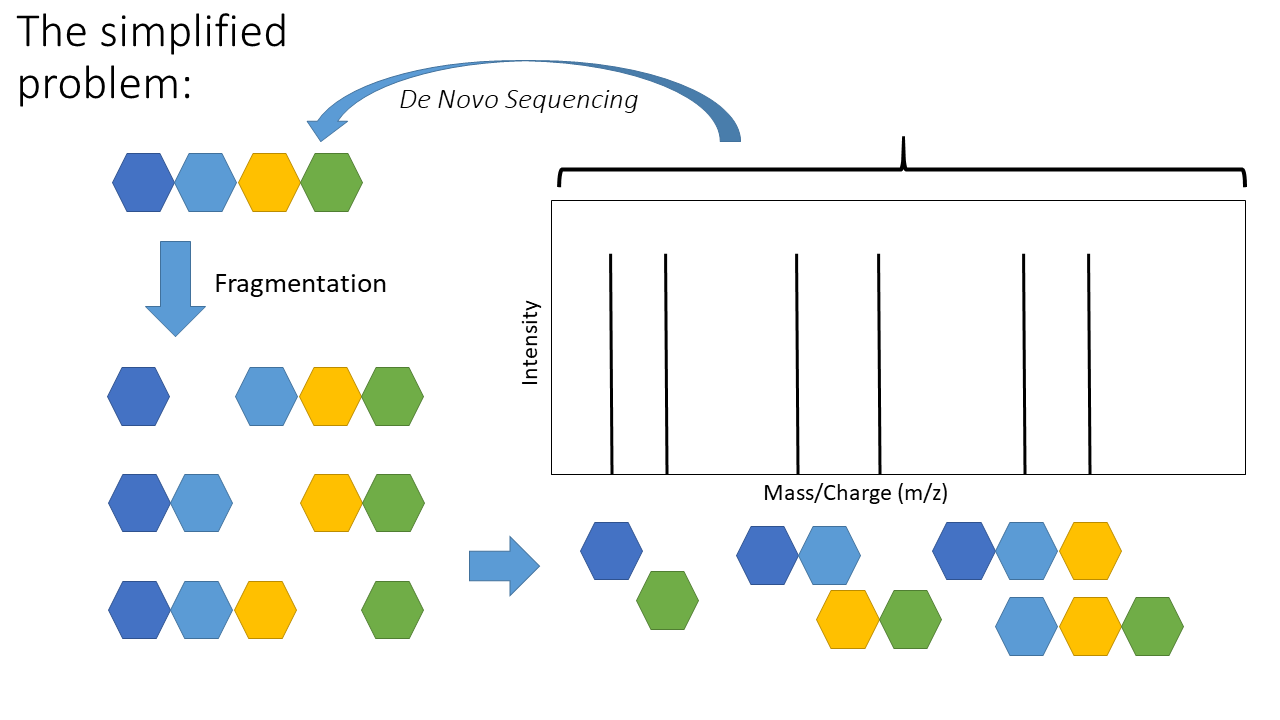
Applied Sciences | Free Full-Text | Denovo-GCN: De Novo Peptide Sequencing by Graph Convolutional Neural Networks
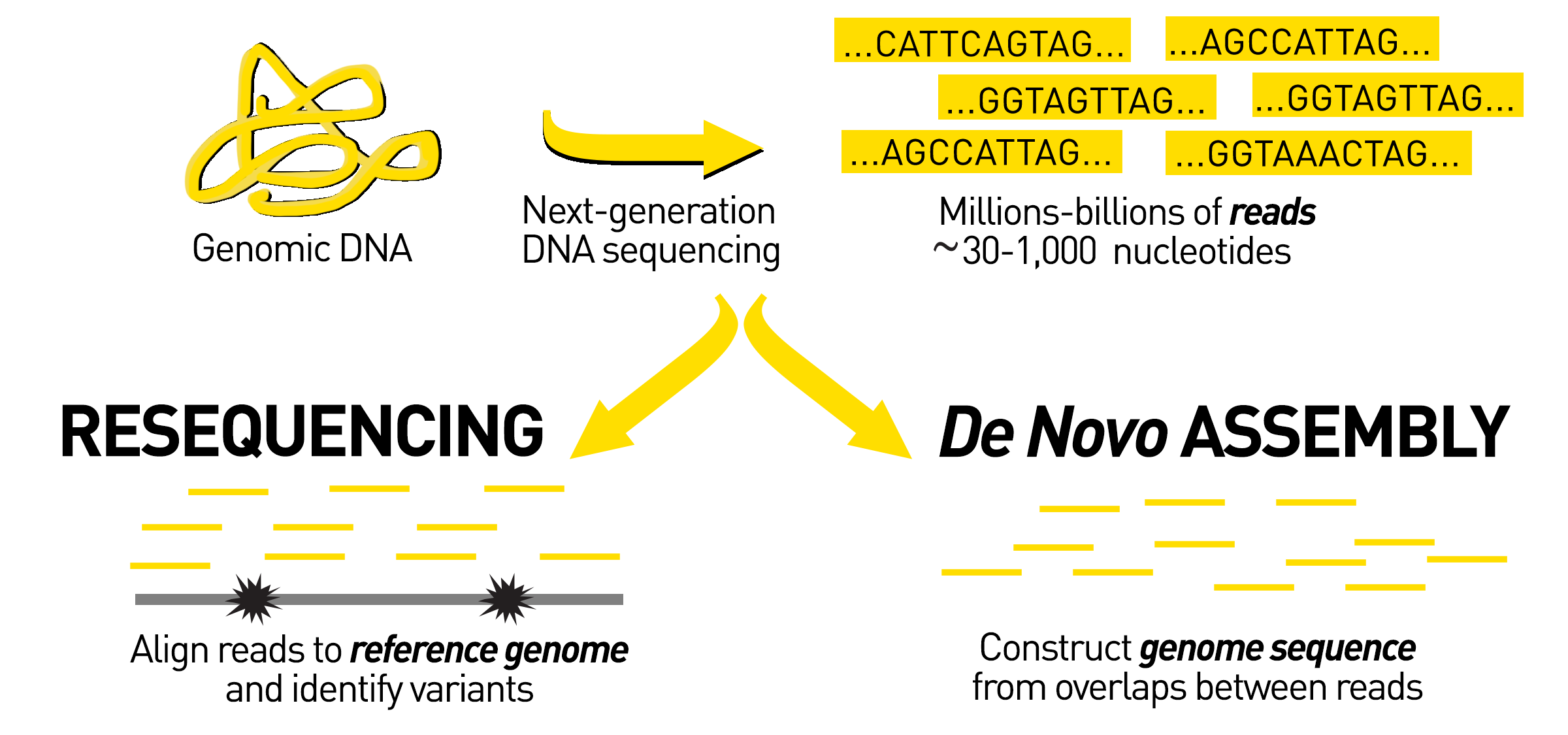
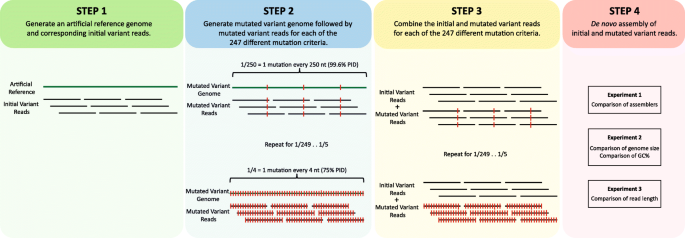
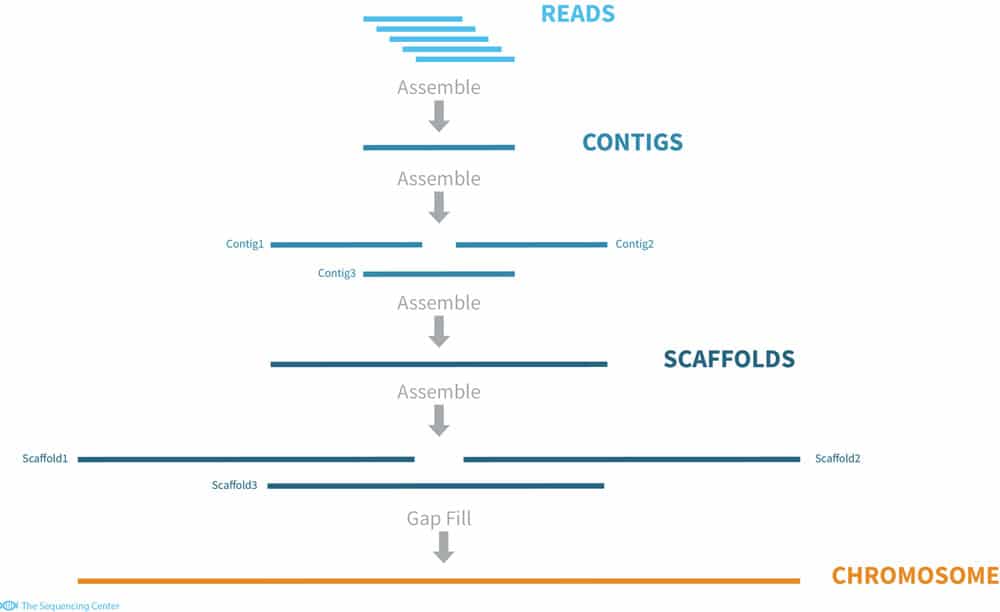
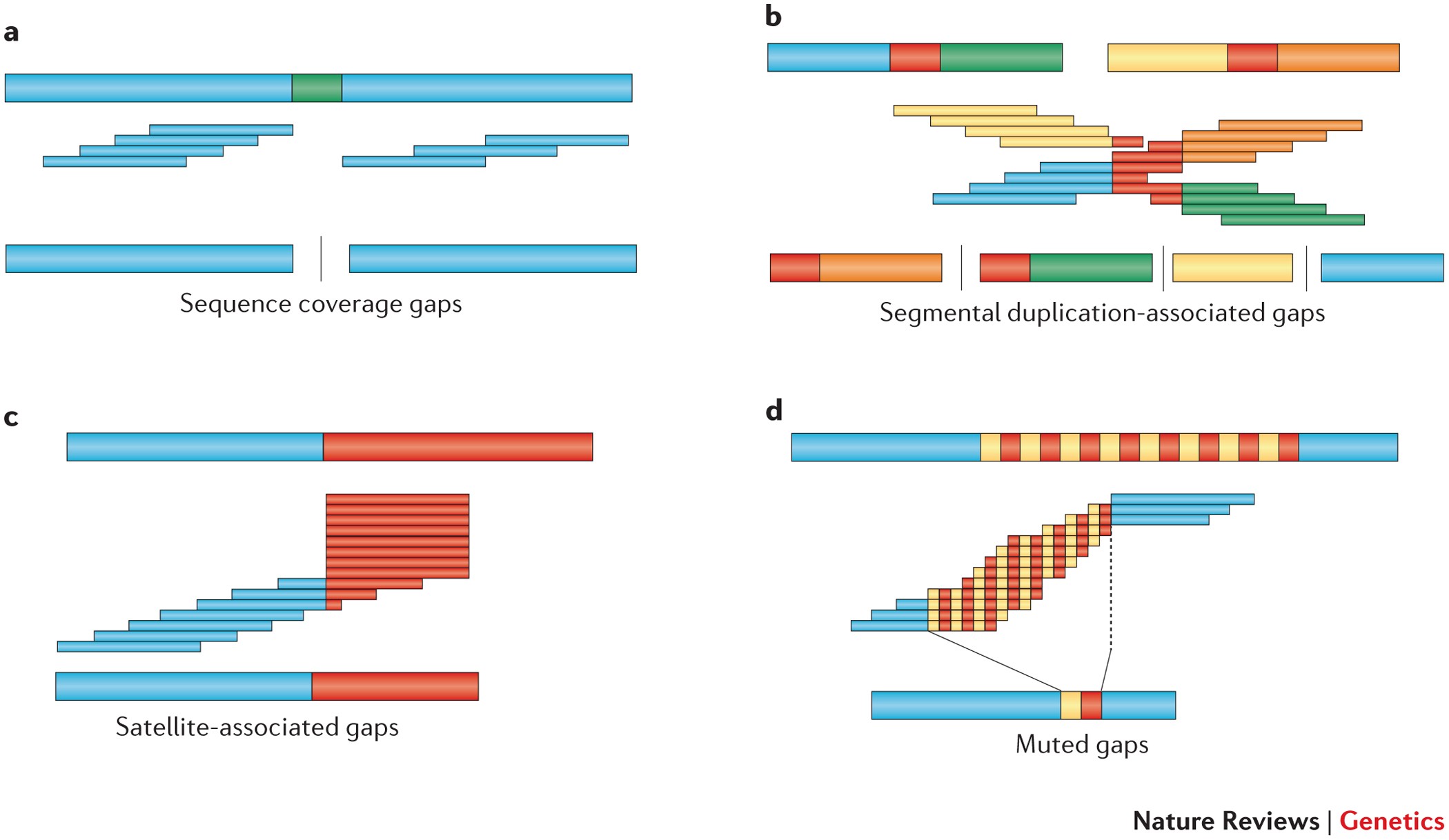
Sequencing from scratch: reference genomes and de novo sequence assembly – HudsonAlpha Institute for Biotechnology
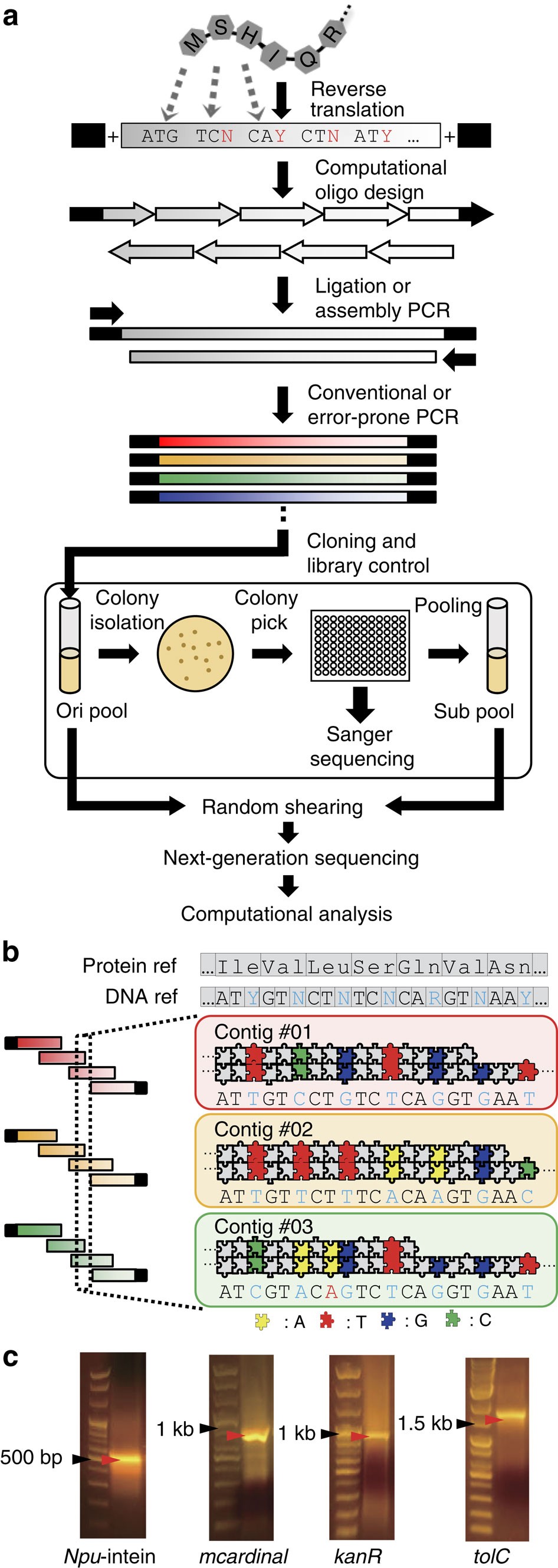
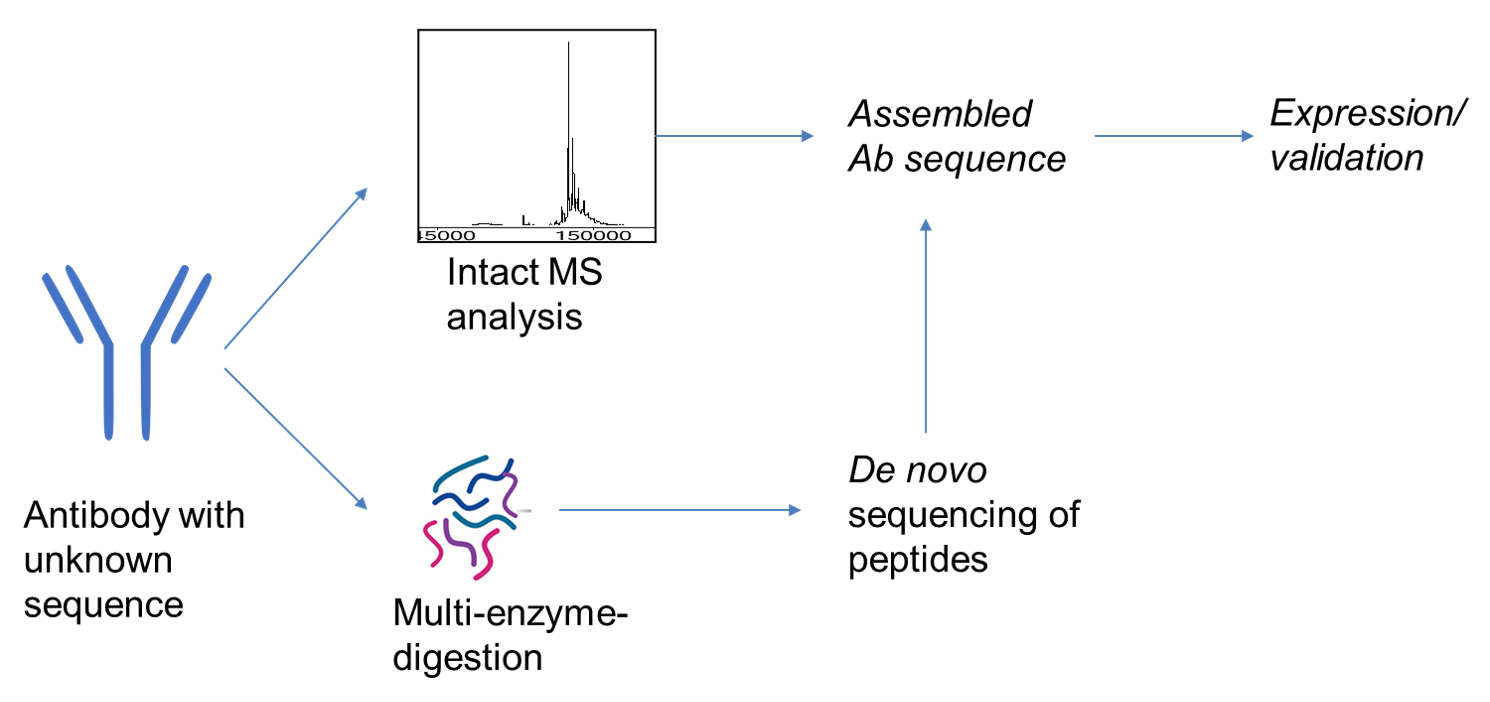
De novo assembly and next-generation sequencing to analyse full-length gene variants from codon-barcoded libraries | Nature Communications