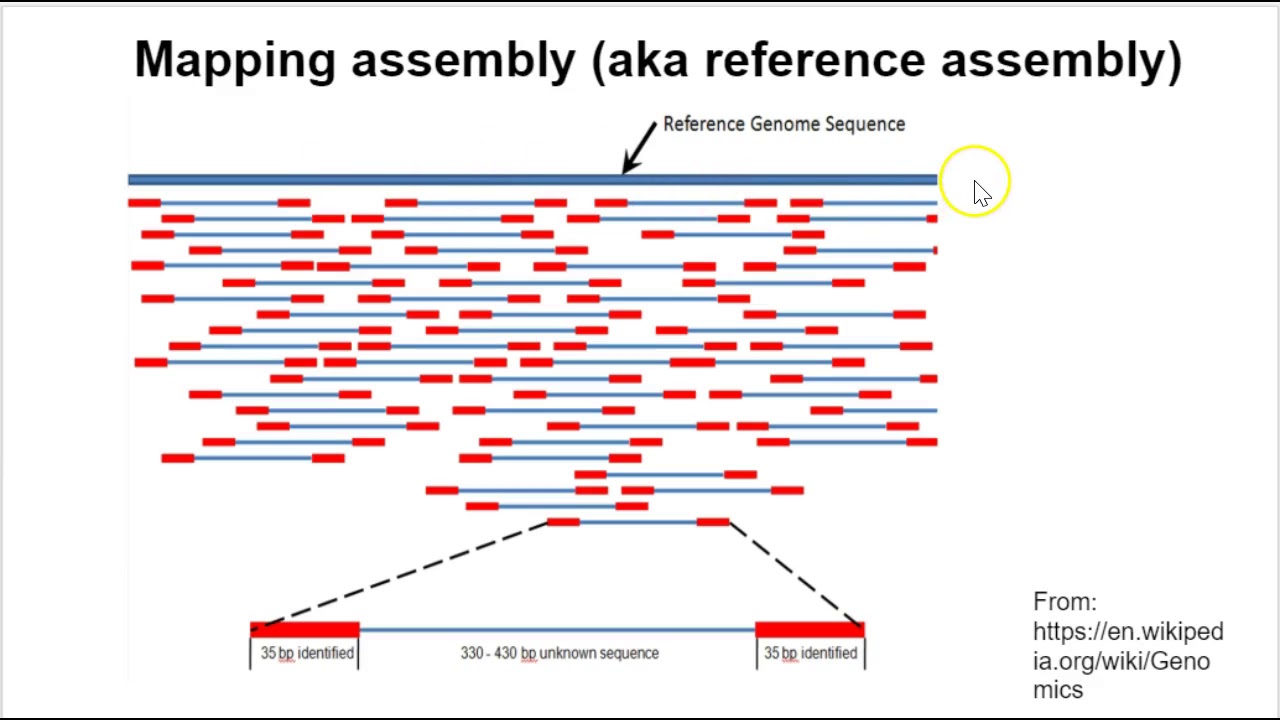
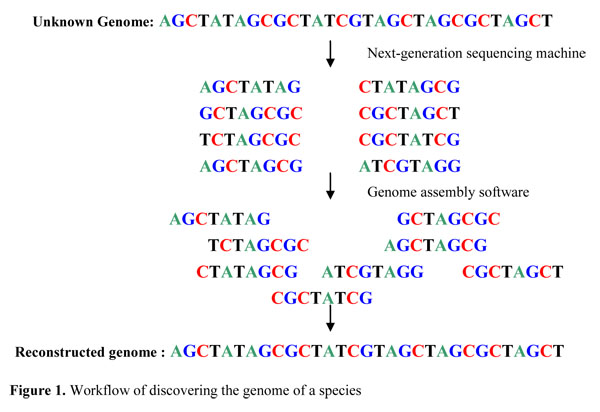
Sequencing from scratch: reference genomes and de novo sequence assembly – HudsonAlpha Institute for Biotechnology
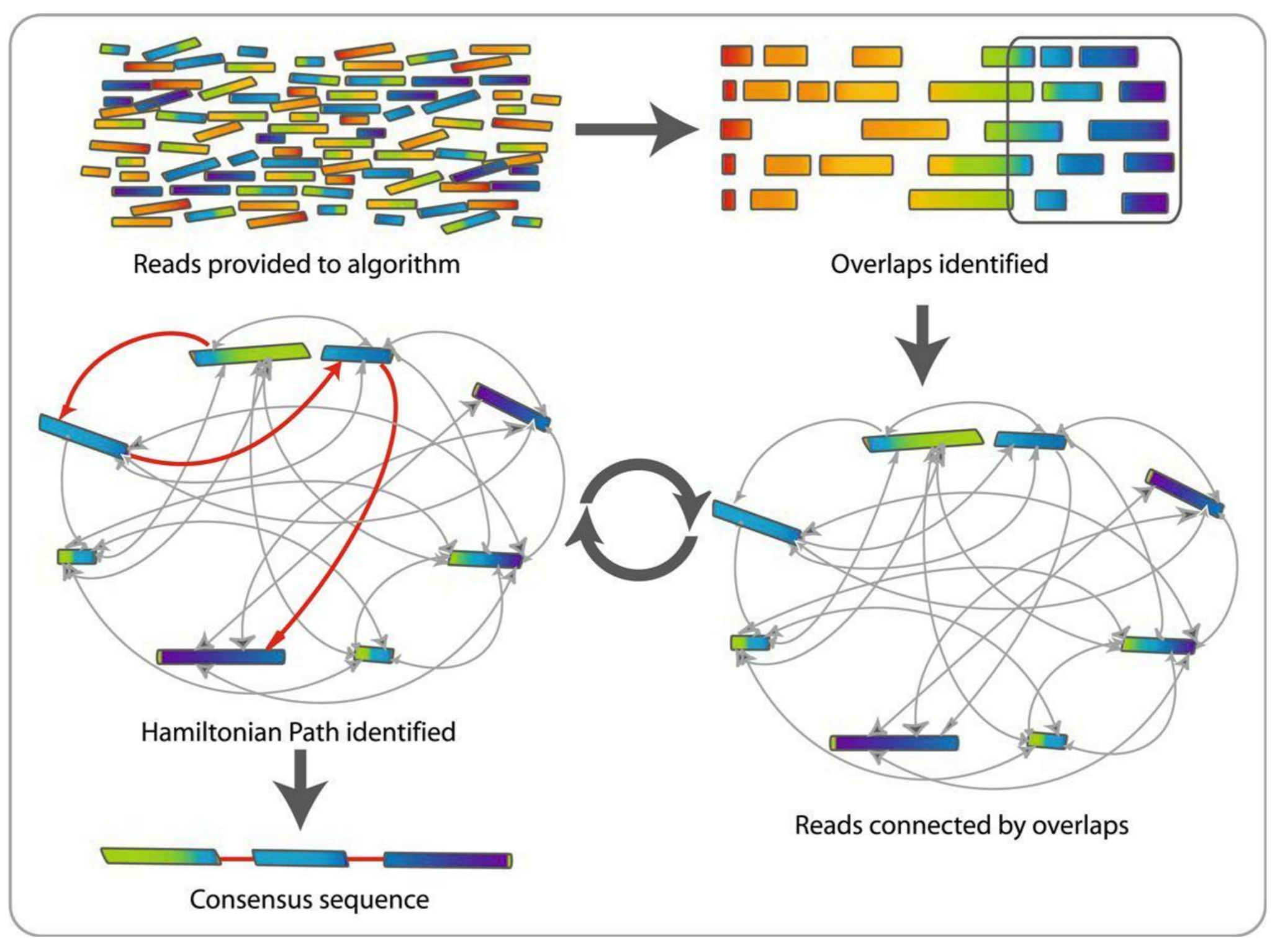
Slide Deck: Slide Deck: Hands-on: Unicycler assembly of SARS-CoV-2 genome with preprocessing to remove human genome reads / Assembly

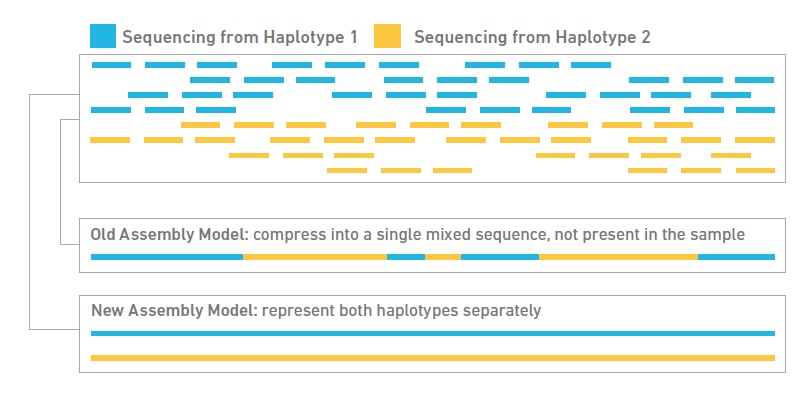
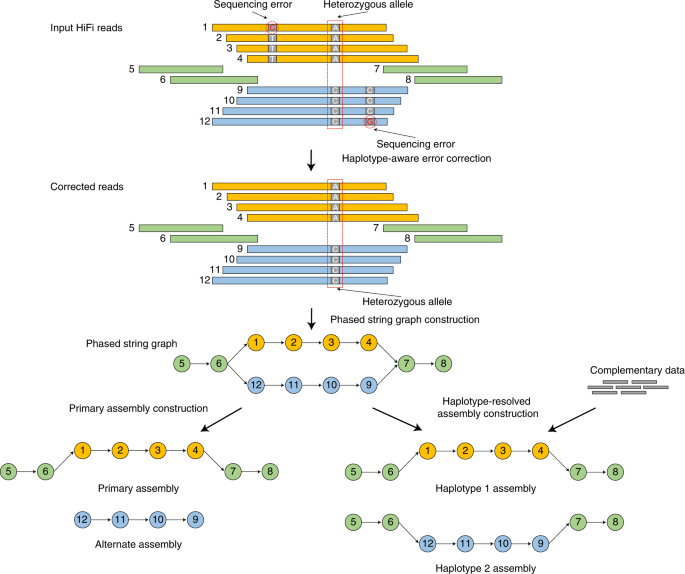
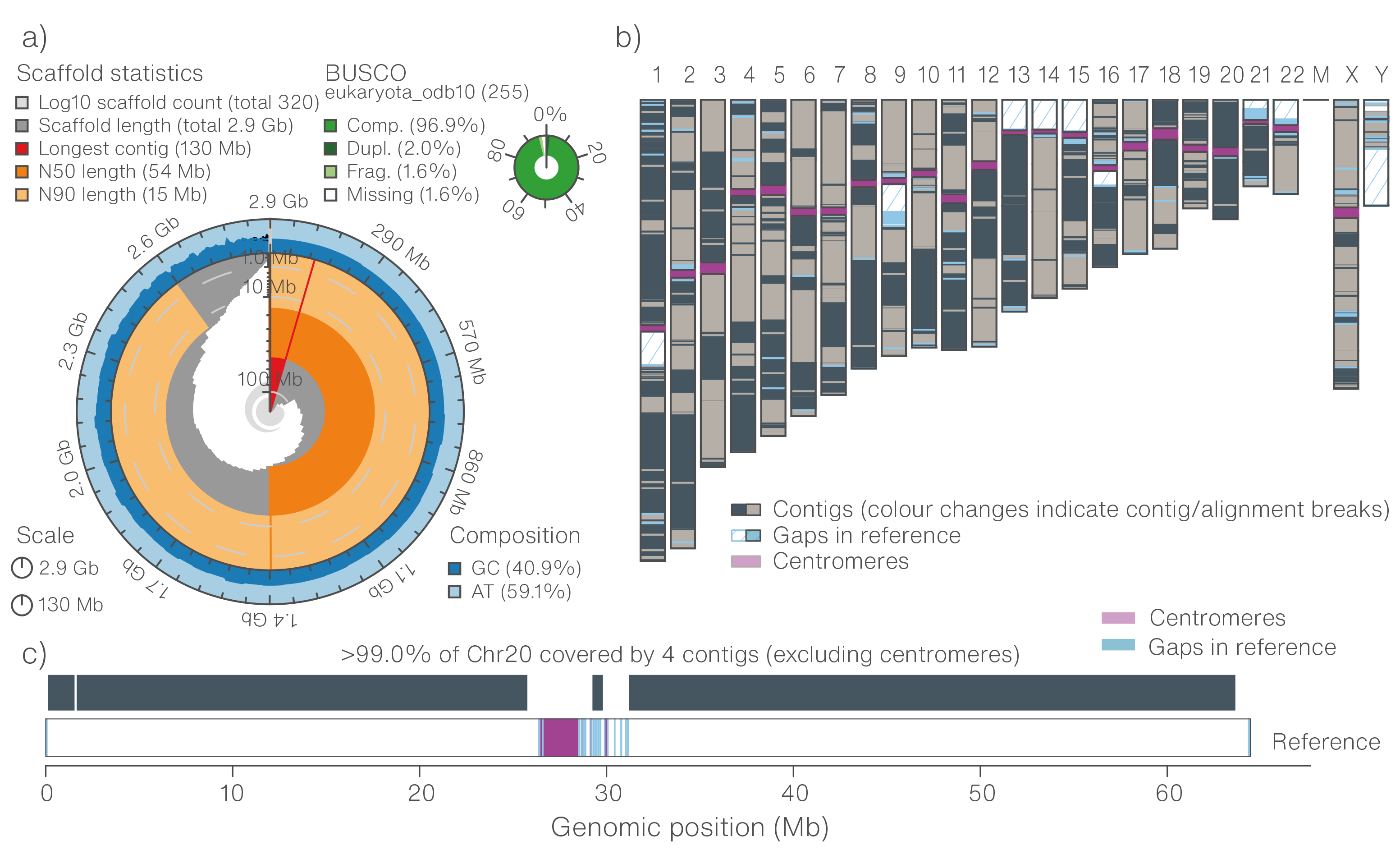
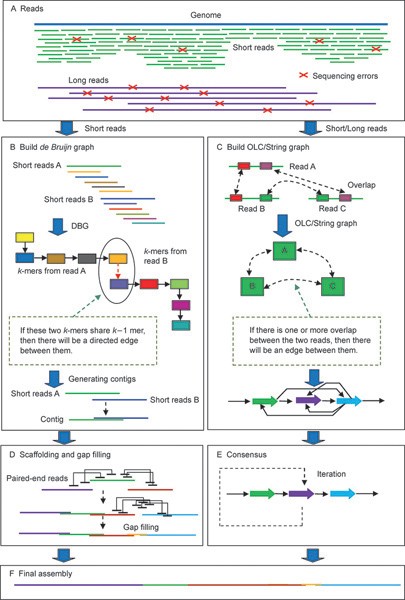
Tools and Strategies for Long-Read Sequencing and De Novo Assembly of Plant Genomes: Trends in Plant Science

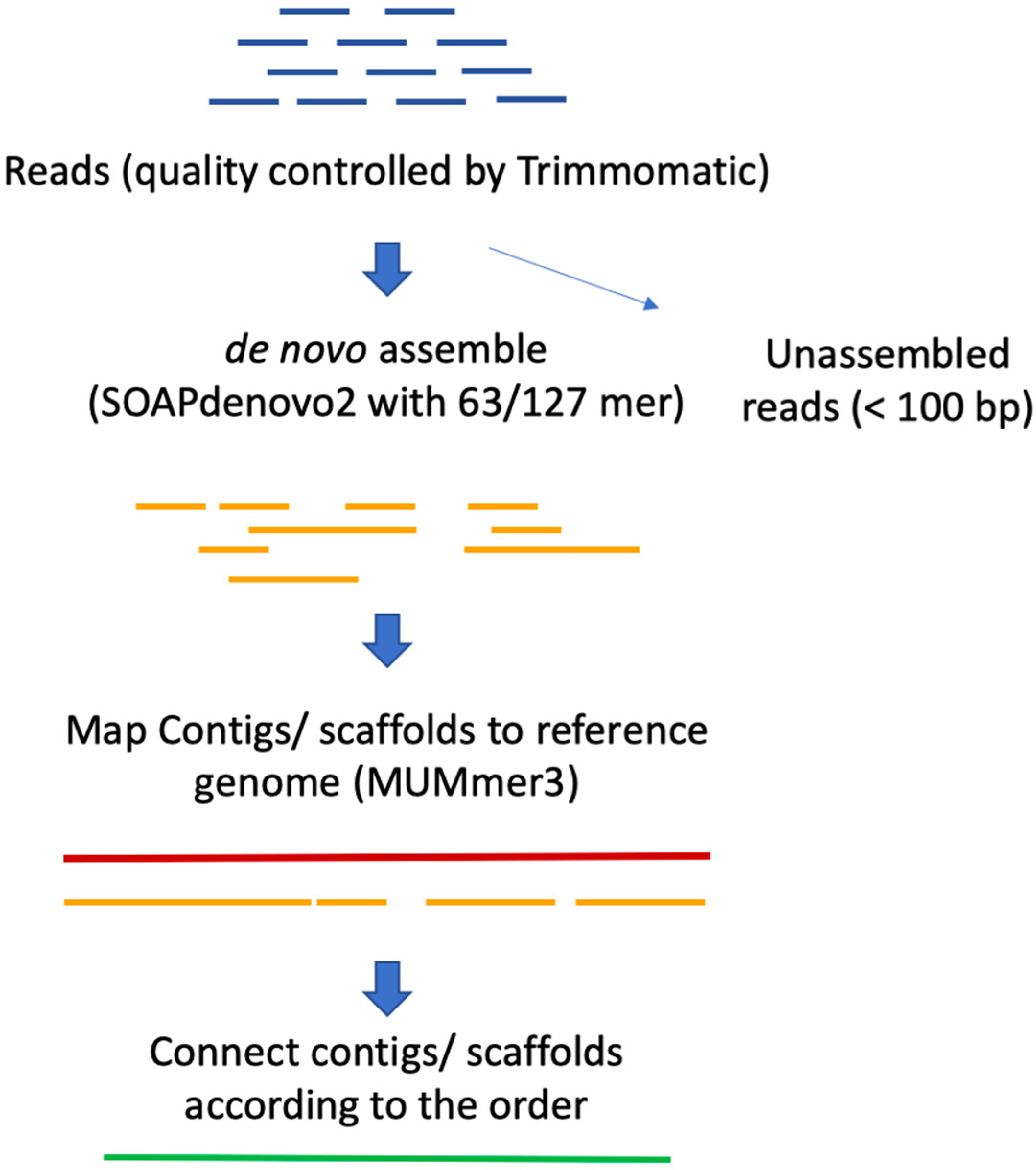
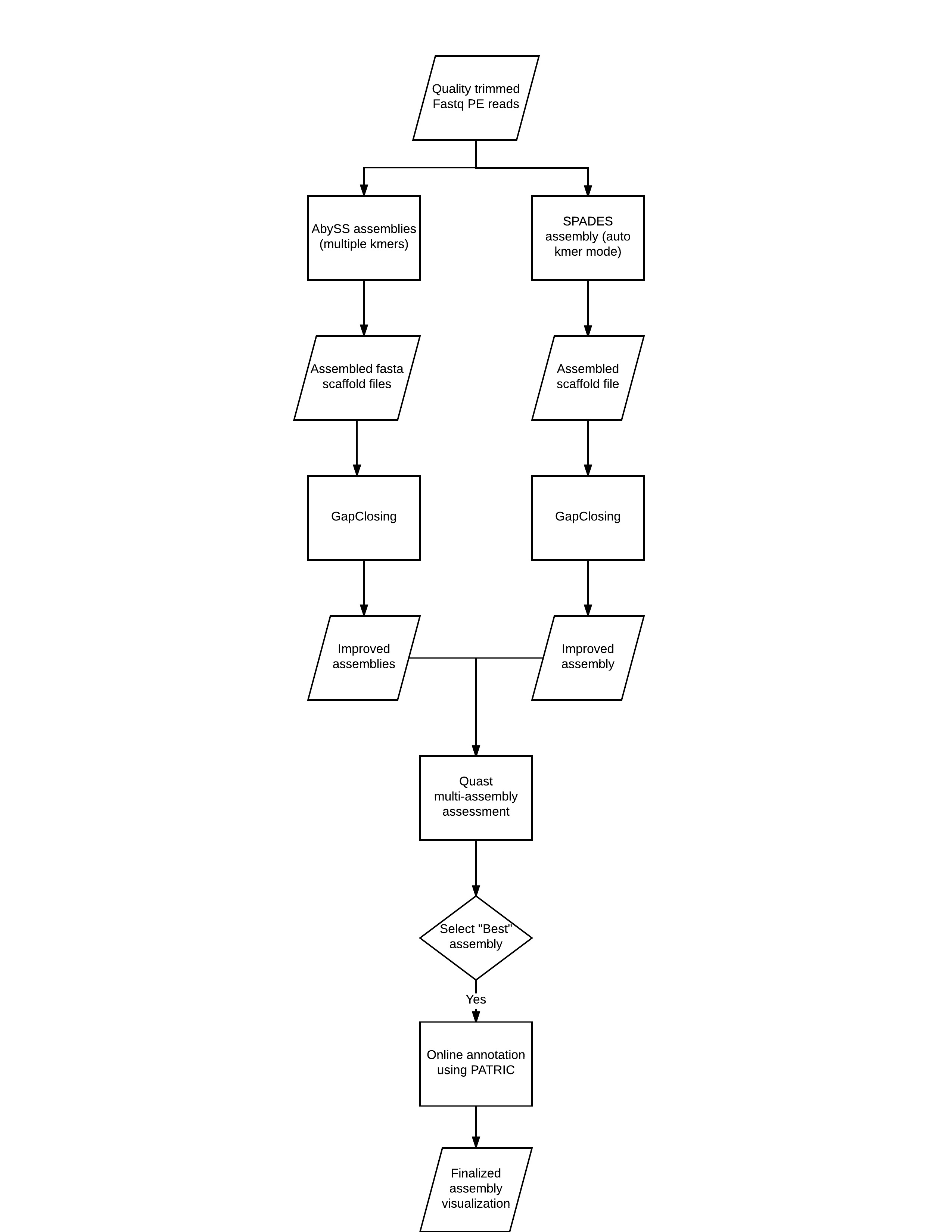
High-throughput pipeline for the de novo viral genome assembly and identification of minority variants from Next-Generation Sequencing of residual diagnostic samples | bioRxiv

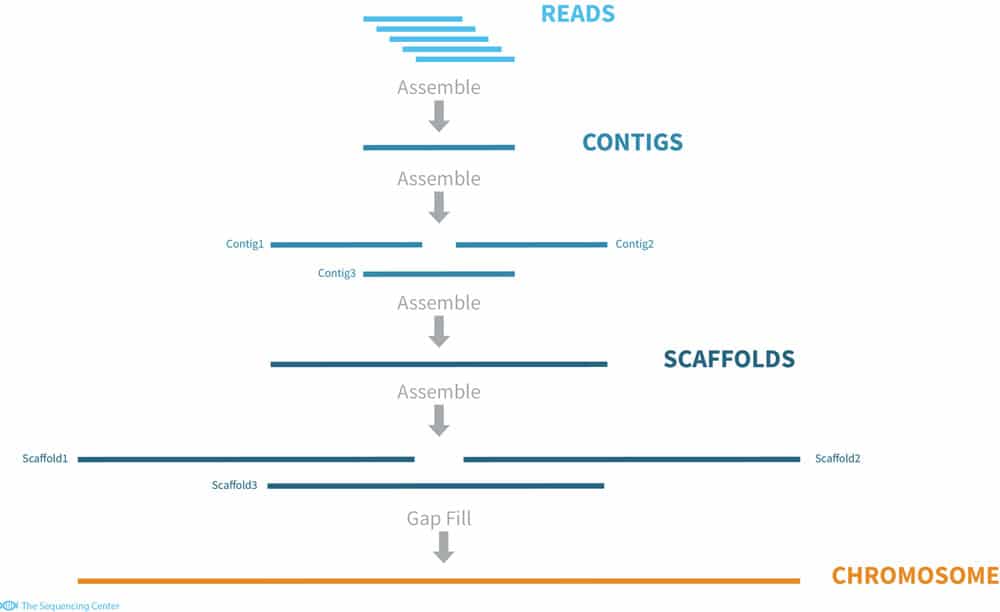
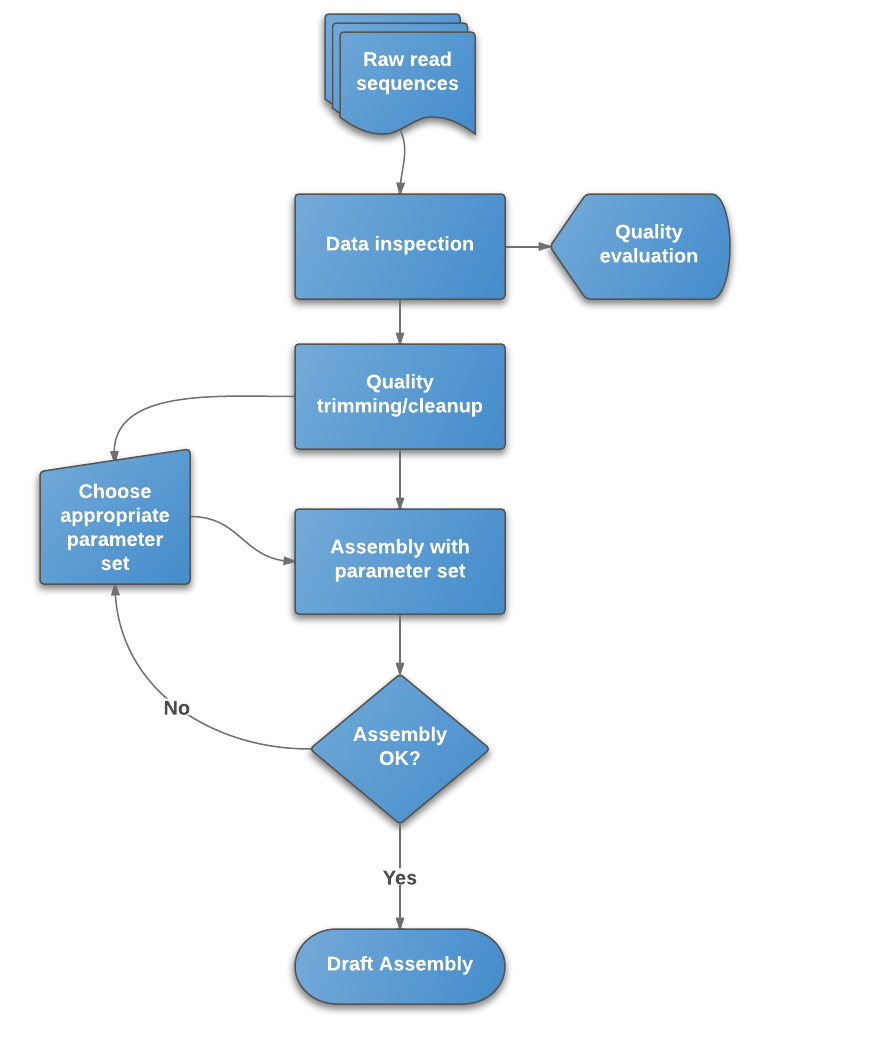
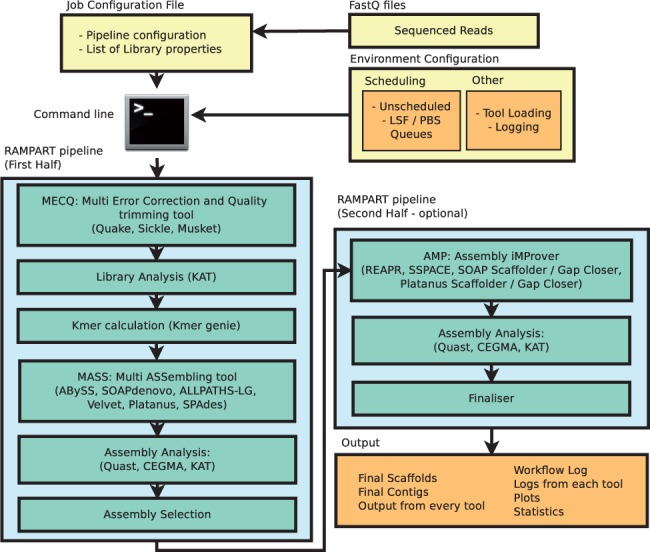
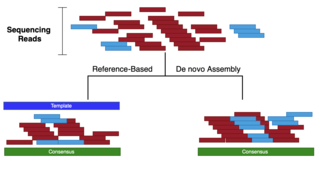
Whole Genome de novo Assembly |Supporting de novo assembly training |Course |WGS sequencing | data analysis| Workshop